GRASS: SEMI-AUTOMATED NMR-BASED STRUCTURE ELUCIDATION OF SACCHARIDES
aHigher Chemical College of the Russian Academy of Sciences, Moscow, Russia
bN.D. Zelinsky Institute of Organic Chemistry, Russian Academy of Sciences, Moscow, Russia
KEYWORDS: NMR simulation, structure prediction, glycoinformatics
Bioinformatics, 2018, v.34(6), pp. 957-963
DOI: 10.1093/bioinformatics/btx696, PMID: 29092007
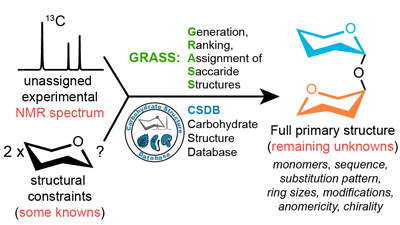
Motivation
Carbohydrates play crucial roles in various biochemical processes and are useful for developing drugs and vaccines. However, in case of carbohydrates, the primary structure elucidation is usually a sophisticated task. Therefore, they remain the least structurally characterized class of biomolecules, and this breach inhibits the progress in glycochemistry and glycobiology. Creating a usable instrument designed to assist researchers in natural carbohydrate structure determination would advance glycochemistry in biomedical and pharmaceutical applications.
Results
We present GRASS (Generation, Ranking and Assignment of Saccharide Structures), a novel method for semi-automated elucidation of carbohydrate and derivative structures which uses unassigned 13C NMR spectra and information obtained from chromatography, optical, chemical and other methods. This approach is based on new methods of carbohydrate NMR simulation recently reported as the most accurate. It combines a broad diversity of supported structural features, high accuracy and performance.
Availability
GRASS is impelmented in a free web tool available at http://csdb.glycoscience.ru/grass.html