BIOPSEL : substitution effect database
The substitution effect database is filled in with known data on glycosylation and phosphorylation effects and contains information on about 150 variants of substitution. The way to find the certain effect is demonstrated on the scheme:
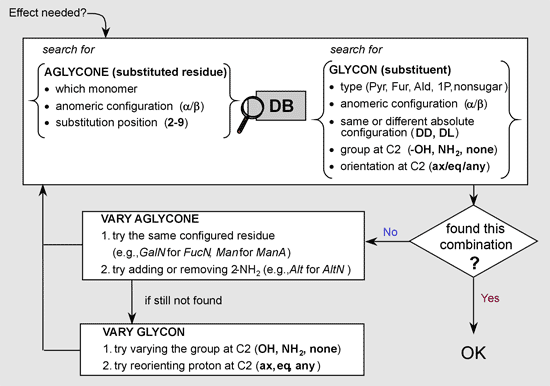
Each entry in the effect database contains the following information:
- properties of the substituted (main) residue :
- which residue (reference to the entry in the residue database)
- its anomeric configuration, if it may have anomeric forms, if not specified, data are used for both anomers
- the substitution position
- properties of the substituent:
- the type of substituent: pyranose, furanose, alditol, non-carbohydrate
- anomeric configuration, if it may have anomeric forms
- the presence or absence of the phosphate group at C1
- additional groups attached to C1 (none, carboxyl group, C-chain)
- the type of substituter at C2 (-OH/-NH2/deoxy)
- the orientation of proton at C2 (axial/equatorial/unoriented or both)
- the combination of absolute configurations of the 1st substituentand the main residue (DD/DL)
If the desired effect is missing from the database, its engine tries 1.) adding or removing amino group at C2 of the substited residue, 2.) searching for another monomer with the same configuration , e.g. GalNAc instead of FucNAc. If these effects are missing too, database engine tries to vary the type and orientation of the group at C2 of the substituent.
The glycosylation effects for three widespread sugar configurations (glc, gal, man) are represented most completely, so the prediction of structure of polymers built of widespread carbohydrate residues only shows the best accuracy. In the case of not more than one non-carbohydrate or rarely-occurring residue per repeating unit, the probability to find the proper structure among 1st five ones generated is about 95%.
Last update: 2002  Home
Home
