TERTIARY STRUCTURE OF BACTERIAL MUREIN: THE SCAFFOLD MODEL
N. F. Gamaleya Institute for Epidemiology and Microbiology, Moscow, Russia,
N. D. Zelinsky Institute of Organic Chemistry, Moscow, Russia,
Department of Immunochemistry and Biochemical Microbiology, Research Center Borstel,
Center for Medicine and Biosciences, Germany
KEYWORDS: tertiary structure, peptidoglycan, bacterial murein, computer simulation, scaffold
Journal of Bacteriology, 2003, v.185(11), pp.3458-3468
DOI: 10.1128/JB.185.11.3458-3468.2003 (free full text)
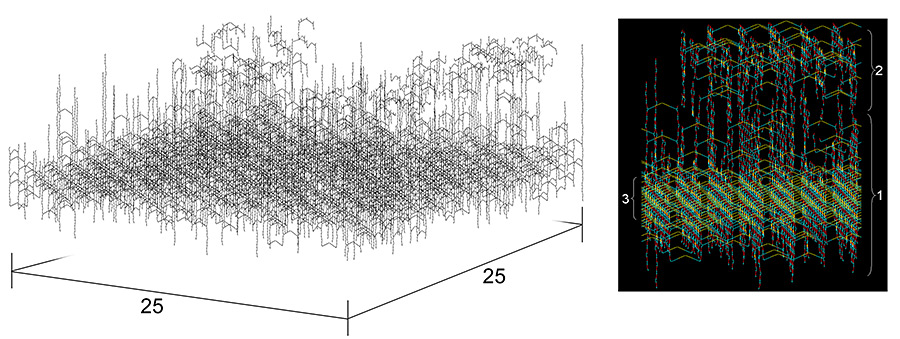
Although the chemical structure and physical properties of peptidoglycan have been elucidated for some time, the precise three-dimensional organization of murein has remained elusive. Earlier published computer simulations of the bacterial murein architecture modeled peptidoglycan strands in either a regular (D. Pink, J. Moeller, B. Quinn, M. Jericho, and T. Beveridge, J. Bacteriol. 182: 5925-5930, 2000) or an irregular (A. Koch, J. Theor. Biol. 204: 533-541, 2000) parallel orientation with respect to the plasma membrane. However, after integrating published experimental data on glycan chain length distribution and the degree of peptide side chain cross-linking into this computer simulation, we now report that the proposed planar network of murein appears largely dysfunctional. In contrast, a scaffold model of murein architecture, which assumes that glycan strands extend perpendicularly to the plasma membrane, was found to accommodate published experimental evidence and yield a viable stress-bearing matrix. Moreover, this model is in accordance with the well-established principle of murein assembly in vivo, i.e., sequential attachment of strands to the preexisting structure. For the first time, the phenomenon of division plane alternation in dividing bacteria can be reconciled with a computer model of the molecular architecture of murein.